The Alam Lab focuses on mechanistic studies of asthma.
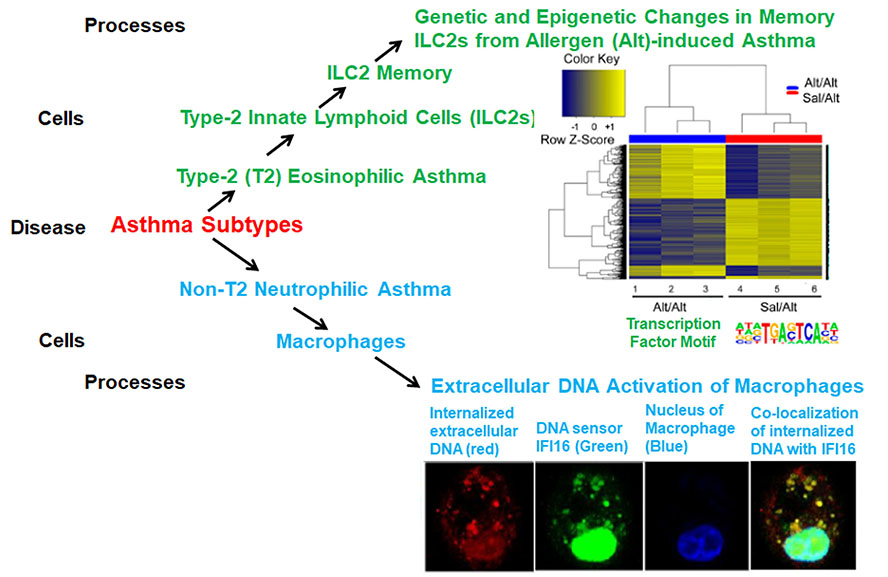
The Alam Lab focuses on mechanistic studies of asthma. We study molecular, signaling and cellular mechanisms of various endotypes of asthma. In one project we examine the role of type-2 innate lymphoid cells (ILC2s) in asthma. We examine ILC2 recognition of allergen-associated molecular patterns (PAMPs), generation of ILC2 memory for PAMPs, the genetic, epigenetic and signaling mechanisms for generation of memory, identification of memory-associated enhancers and their relevance for asthma. The underlying hypothesis is that memory enhancer-driven ILC2 activation is largely responsible for persistence of asthma. We study blood and lung ILC2s from the mouse model of asthma and from asthmatic patients. Human memory ILC2s are relatively steroid-resistant. We study the molecular mechanism of steroid resistance of ILC2s and Th2 cells obtained from asthma patients. The foregoing studies are designed to delineate the role of ILC2s in type-2 (T-2) asthma. In another project we study non-T2 neutrophilic asthma. We examine the role of host-derived extracellular DNA on activation of cellular DNA sensors and induction of neutrophilic asthma. We study the mechanism of DNA internalization by airway macrophages, the activation of the DNA sensor IFI16 and the STING signaling pathway, the induction of neutrophilic inflammation, and the role of allergens and the IL33-ST2 axis in the foregoing processes. We apply state-of-art technologies (scRNA-seq, ATAC-seq, RNA-seq, ChIP, CRISPR-Cas9 gene editing, knockout and mutant mouse strains, flow cytometry and mass spectrometry) to animal models and clinical samples in order to generate fundamental knowledge about various endotypes of asthma, and identify novel therapeutic targets. Once we identify key molecules and pathways, we target them with pharmacological agents and biologics in preclinical models.

Rafeul Alam, MD, PhD
Lab Resources & Services
The Lab has unique expertise in several areas:
Disease Models: Mouse models of asthma: Acute asthma, chronic asthma, ILC2-driven asthma, neutrophilic asthma, steroid-resistant asthma, humanized model of asthma
- Methodologies: Flow cytometry and genomic-based immunophenotyping (ILC2, Th2, Th2/Th17 and neutrophilic) of asthma using tissues samples (lung, blood, bronchoalveolar lavage), scRNA-seq, RNA-seq, ATAC-seq, ChIP, CRISPR-Cas9 mediated gene editing, and measurement of lung function on live mice by FlexiVent
Current Projects
Research Theme
Lab Personnel
Lab Staff
Anand Sripada, PhD
Kapil Sirohi, PhD
Rangati Varma, PhD
Anita Sahu, PhD
Carolyn Kwiat, MD
Lakshmi Gopalakrishnan Nair, MD
Neetigyata Pratap Singh, MS
Ka Ho Cheung, MD
Research Coordinator
Allen Stevens, BA
IRB Coordinator
Christena Kolakowski, MS
Collaborators
Magdalena Gorska, MD, PhD
Richard J. Martin, MD
HongWei Chu, MD, PhD
Laurie Manka, MD
Vamsi Guntur, MD
Michael Wechsler, MD
Anthony Gerber, MD
Arnav Gupta, MD
Camille Moore, PhD
Brian Vestal, PhD
Jessica Galant-Swafford, MD
Administrative Assistant
Lien Hang, BA/BS
Recent Publications
- Liu S, Sirohi K, Verma M, McKay J, Michalec L, Sripada A, Danhorn T, Rollins D, Good J, Martin RJ, Gorska MM and Alam R. Optimal identification of human conventional and nonconventional (CRTH2-IL7Rα-) ILC2s using additional surface markers. J Allergy Clin Immunol. 2020;146(2):390-405. doi:10.1016/j.jaci.2020.01.038
- Sripada A, Sirohi K, Michalec L, Gorentla B, Guo L, McKay J, Yadav S, Verma M, Good J, Rollins D, Gorska MM and Alam R. Sprouty2 positively regulates T cell function and airway inflammation through regulation of CSK and LCK. PLoS Biol. 2021 Mar 8;19(3):e3001063. doi: 10.1371/journal.pbio.3001063. PMID: 33684096
- Verma M, Michalec L, Sripada A, McKay J, Sirohi K, Verma D, Sheth D, Martin R, Dyjack N, Seibold MA, Knapp JR, Tu TH, O'Connor BP, Gorska MM, Alam R. The molecular and epigenetic mechanisms of innate lymphoid cell (ILC) memory and its relevance for asthma. J Exp Med. 2021 Jul 5;218(7):e20201354. doi: 10.1084/jem.20201354. Epub 2021 Jun 2. PMID: 34076685; PMCID: PMC8176441.
- Sripada A, Verma D, Varma R, Sirohi K, Kwiat C, Pathria M, Verma M, Sahu A, Guntur VP, Manka LA, Vestal B, Moore CM, Martin RJ, Gorska MM, Cambier J, Getahun A, Alam R. Allergens abrogate anti-inflammatory DNA effects and unmasks macrophage-driven neutrophilic asthma via ILC2/STING/TNFa signaling. J Clin Invest. 2025 Jun 17:e187907. doi: 10.1172/JCI187907. Epub ahead of print. PMID: 40526428.