Comparative Genomics & Innate Immunity
The Innate Immune Response and Human Disease
The human immune response has two components: a rapid (innate) response and a slower but more specific (adaptive) response. The adaptive immune system involves well characterized cells and immune mediators such as B cells and antibodies. The innate immune response is less well understood but involves the action of phagocytic and cytotoxic cells that migrate to infection sites and fight the pathogen. Over the past decade, it has become clear that the innate immune system plays a critical role in the pathogenesis of many human diseases, ranging from sepsis to asthma to cancer to atherosclerosis. Thus, the identification of genes that regulate the body's innate immune system can have profound consequences for the diagnosis and treatment of many very common and devastating diseases.
Identifying Innate Immune Regulators
Numerous genetic and biochemical approaches have been taken to identify innate immune regulators. With the advent of new genomic technologies, it is now possible to take a global approach to investigate innate immune gene function on a whole-genome scale. Such approaches are powerful but also raise several critical issues. How can we efficiently sift through the many genes identified in genomic approaches to find the critical immune and disease regulators? How can we determine which of these genes will play a role in human immunity and disease? In many cases, genes identified in disease models in model organisms fail to function similarly when tested in humans.
Comparative Genomics
To maximize our chances of identifying genes that affect human disease, we are taking a comparative genomics approach to identify and investigate innate immune regulators (Figure 1). Comparative genomics is the comparison of gene sequence and function across species, which can lead to a better understanding of disease.
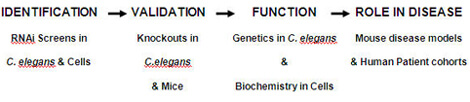
We are using model systems such as the nematode C. elegans and mouse macrophage cell culture to identify innate immune regulators. These relatively simple model systems are readily amenable to investigation. For example, we can use RNA-interference (RNAi) to inhibit all genes in the genome in these systems in rapid fashion, a powerful technique that is currently impossible in higher organisms.
Our hypothesis is that genes that affect innate immunity in more than one of these relatively simple model systems are exhibiting evolutionarily conserved function. Therefore, these conserved innate immune genes are likely to regulate immunity and disease in mammals.
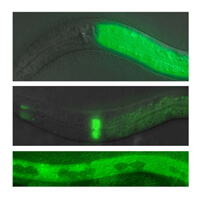
We are further investigating candidate genes identified in these simple models using knockout mutants in C. elegans and mice. These knockout mutants are used to validate the RNAi results and are also used to investigate how these novel genes affect the immune system.
Finally, we are also examining DNA polymorphisms identified in our candidate genes in human patient cohorts to see if these polymorphisms alter the risk of human disease. By identifying such disease-associated DNA polymorphisms, we can offer clinicians a potentially rich resource to assist in diagnosis and disease treatment using a personalized approach to medicine.
